The following is a data model and basic implementation plan for an online animal classification system. This system contains multimedia records for many types of animal. The system allows for the construction of a variety of classifications by using Node objects. By adding new classifications or exploring existing ones, users can examine a variety of relationships between animals. They can browse alternate classifications, or even intersect different classifications in searches, depending on their current information need.
I imagine this system as a web-based, open-access system. It will be an animal encyclopedia with text and images of specific animal species. It can serve as a partial index to related works, especially since cited resources can be URLs as well as print materials. It will be a taxonomy modeling tool. Most importantly, the combination of multiple user-created classifications will make it a powerful searching and meta-data system.
The system would probably be setup and maintained by either a volunteer group, non-profit organization, or university. There will be a graphical Web interface so that users can browse, see classification structures, and get the full benefit of the images and URL links to other sources. The backend database will be implemented using PostgreSQL.

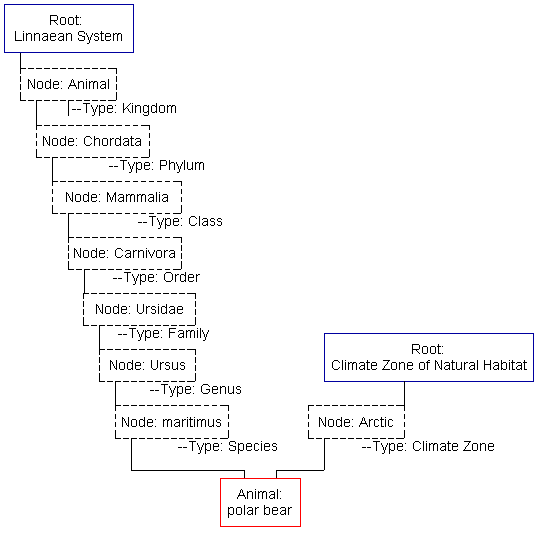
This image shows the three major elements of this system. The animal record (here a polar bear) will contain encyclopedia-like information: an image, text description of the characteristics, habitat, behavior, etc., and citations to further resources. Animal records can exist in any number of classifications, even none at all.
Roots are the tops of a user-created classification scheme. They contain a short description of the classification and links to the top level of contained nodes.
Nodes form the tree-like structure of a classification. (In this image, only one "branch" of each classification tree is shown.) Classifications may only have one layer of nodes, such as the "Climate" classification in this example. Other nodes under this root might be "Tundra", "Rain Forest", "Desert", "Marine", etc. Classifications may also have a multiple levels, as does the "Linnaean" classification in this example. At each level shown here, there are other nodes not shown. For example, under the Ursus node, there are many other species nodes which correspond to the many different species of bear--grizzly, brown, black, etc.
Possible users include:
Users such as high school students and some amateur enthusiasts will likely use the system mainly as an encyclopedia to find more information on certain animals. Such uses do not involve building new classifications or adding animal records. These users will be Users in terms of the SSM below.
Other amateur enthusiasts may compile information from various sources and create new animal records or edit current ones. Taxonomists may use the system to build models of their work. Biology researchers may add their findings to animal records. They may also use the system to examine possible relationships made visible through intersecting classificiations, such as effect of climate on gestation period. These users that create new records will be Creators in terms of the SSM below.
Here are some example information needs that illustrate use of the system.
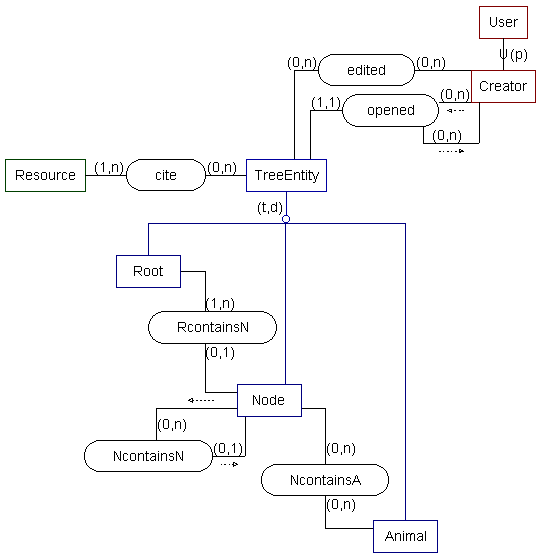
-- SSM model --
| Animal | |
|---|---|
| id | <integer> -- a TreeEntitiy foreign key |
| species_name | <char(60)> -- the Latin Genus-species name for this animal |
| common_name | <char(60)> -- the common English name for this animal. |
| keywords | <varchar(200)> -- keywords describing this animal to aid in searching |
| description | <text> |
| image | filename: <char(30)> -- the name of the image file pixel-width: <integer> -- the width of the image in pixels pixel-height: <integer> -- the height of the image in pixels format: <char(4)> -- {JPEG | GIF | PNG | BMP} comments: <varchar(500)> -- any comments on the image image: <image> |
| Root | |
|---|---|
| id | <integer> -- a TreeEntitiy foreign key |
| classif_name | <char(60)> -- the name of the classification system stemming from this root |
| keywords | <varchar(200)> -- keywords describing this classification to aid in searching |
| scope_all | <boolean> -- whether this classification aims to include all animals (as opposed to just a subset) |
| description | <varchar(2000)> -- a description of this classification system |
| Node | |
|---|---|
| id | <integer> -- a TreeEntitiy foreign key |
| taxon_name | <char(60)> -- the name of the particular taxon or group or classification level represented by this node |
| type | <char(60)> -- describes the type of taxon or the level within the classification system |
| keywords | <varchar(200)> -- keywords describing this node to aid in searching |
| description | <varchar(2000)> -- a description of this entity |
| TreeEntity | |
|---|---|
| id | <sequence> -- a unique integer system id for this entity |
| table_ref | <char(1)> -- {r|n|a} which table to look to to find this entity |
| Resource | |
|---|---|
| id | <integer> |
| author | <char(100)> |
| title | <char(500)> |
| publisher | <char(300)> -- or contain journal title and issue info |
| location | <char(150)> -- or may contain a URL |
| pub_date | <date> |
| notes | <varchar(750)> -- notes or comments about this resource |
| User | |
|---|---|
| username | <char(20)> -- a unique username |
| age | <integer> |
| education | <char(11)> {ELEMENTARY | MIDDLE | HIGH SCHOOL | COLLEGE | GRAD | TEACHER | PROFESSOR} |
| nationality | <char(30)> |
| <char(30)> | |
| created | <date> |
| last_accessed | <date> |
| time_inactive | <derived> -- how long it has been since this user logged in |
| Creator | |
|---|---|
| password | <char(12)> |
| name | <char(50)> |
| address | <char(120)> |
| release_prefs | <char(8)>{USERNAME | NAME | EMAIL | ADDRESS} -- how much information to release to other users; each choice includes those before it. |
| greeting | <vchar(750)> -- an introductory message to other users of the system |
| RcontainsN | |
|---|---|
| E1: Root | (1,n) -- A Root must contain some Nodes. |
| E2: Node | (0,1) -- A Node may or may not be contained by a Root |
| NcontainsN | |
|---|---|
| E1: Node | (0,n) -- A Node may or may not contain other Nodes. |
| E2: Node | (0,1) -- A Node may be contained by another Node. |
| NcontainsA | |
|---|---|
| E1: Node | (0,n) -- A Node may or may not contain Animals |
| E2: Animal | (0,n) -- An Animal may be contained by any number of Nodes, or none at all. |
| cite | |
|---|---|
| E1: TreeEntity | (0,n) -- An entity may cite any number of Resources. |
| E2: Resource | (1,n) -- A Resource must be cited at least once. |
| edited | |
|---|---|
| E1: Creator | (0,n) -- A Creator may edit any number of entities. |
| E2: TreeEntity | (0,n) -- An entity can be edited any number of times. |
| time | <timestamp> -- The time and date the new TreeEntity was edited. |
| opened | |
|---|---|
| E1: Creator | (0,n) -- A creator may not have created an entity yet; if he has, he can create any number of them |
| E2: TreeEntity | (1,1) -- Each set of access permissions is related to only a single Entity. |
| E3: Creators | (0,n) -- A Creator grants access to any number of Creators, 0 being all. |
| time | <timestamp> -- The time and date the new TreeEntity was added. |
I foresee the need to create a table for each SSM entity and relationships above. Thus, I will have the following tables:
tree_entities
roots
nodes
animals
resources
users
creators
cited
edited
opened
r_contains_n
n_contains_n
n_contains_a
Here are some example CREATE statements from this system.
CREATE SEQUENCE teid_counter;
CREATE TABLE tree_entities (
teid INTEGER DEFAULT nextval('teid_counter') PRIMARY KEY,
table_ref char(1)
);
CREATE TABLE roots (
id REFERENCES tree_entities PRIMARY KEY,
classif_name char(60),
keywords varchar(200),
scope_all boolean,
description varchar(2000)
);
CREATE TABLE r_contains_n (
root_id REFERENCES roots,
node_id REFERENCES nodes UNIQUE
);
I plan to index the following columns for faster retrieval (in order of importance):
The "contains" relationships will be used frequently in order to request the next layer of nodes in a classification or retrieve the animals from leaf nodes. NcontainsA.child is indexed to facilitate searching for all nodes that contain this animal. (From this, one could work further up the respective classification trees to determine all the roots that contain the animal.) RcontainsN.child and NcontainsN.child (not in the list above) will likely be indexed by the system for the purpose of maintaining uniqueness. That is, a node can only be contained (be a child) once. As such, there should also be a constraint such that a node cannot appear in both NcontainsN.child and RcontainsN.child at the same time.
If possible, it would be nice to also form indices of keywords from the content of all the keywords and descriptions fields, as well as the animal text objects. This would require an IR indexing function, which I currently do not have. It is debatable whether the expense of creating one (if there is not one available from another source) would outweigh the extra system time to run without keyword indexes. Once the database is fully populated, there may be around one million animal records, each with a text object. Performing a sequential text search through all of these would be a heavy undertaking for the system. Yet, as discussed below, there is the possibility of allowing regular expressions searches, which implies necessarily searching against the full text and not an index. In addition, by the time the system is fully populated and text searches become so unwieldy, there should be enough classifications to allow some search area restrictions. I propose that, if the user does not restrict her search to a certain root, node, or intersection, the system should search against an index of keywords. If the user does restrict their domain (and so reduce the cost of the search), then regular expressions and searching against the original text would be allowed.
Certain functions will also be required. They should do the following:
get_contained_nodes(TEid)
get_leaf_nodes(TEid)
get_contained_animals(TEid))
get_all_contained_nodes(TEid)
get_contained_animals(TEid)
These functions are examined in more detail below. Note that all are currently written only to handle nodes. All need to be extended to also accept root TEids, but the process will be the same. These functions are basically elaborate (or not so elaborate) select clauses and so do not require much optimization--they need not be performed before joins. Because the database content changes over time, the results cannot be cached.
This function is not necessary, but, due the frequency of use, it is convenient.
CREATE FUNCTION get_contained_nodes (integer) RETURNS setof integer AS'
SELECT child FROM NcontainsN WHERE parent = $1;
' language 'sql';
After hours of work, I was unable to construct this function using PL/pgSQL, which is PostgreSQL's default procedural language. I was unable to create a table (temporary or otherwise) nor instantiate an array from within a function. This made looping over a group of nodes impossible. While there is a FOR loop in PL/pgSQL that will iterate over the results of a SELECT clause, the results must be in sequence, which node ids are not. Due to time constraints and minimal Linux skills, I was unable to install the PL/Perl module. Thus, this function is represented here only in pseudo-code. This function would be easiest written in in a language with flexible arrays (not of fixed size), such as Perl.
function get_leaf_nodes(integer) {
child_nodes[] = get_contained_nodes($1);
leaf_nodes[];
if (child_nodes.size == 0){
//then this is a leaf node; return the current teid
return $1; //as array of one element, if necessary
}else{
foreach node in child_nodes[]{
//return the leaf nodes in each subtree
leaf_nodes[] = leaf_nodes[] + get_leaf_nodes($1);
}
return leaf_nodes[]
}
}//end function
This function would be nearly identical to get_leaf_nodes. The only difference is that the line
leaf_nodes[] = leaf_nodes[] + get_leaf_nodes($1);
should instead read
leaf_nodes[] = leaf_nodes[] + $1 + get_leaf_nodes($1);
This ensures that every node is added to the array as the function recurses through the tree.
The usefulness of this function is not, at this time, glaringly obvious. But it may be useful in rendering the web interface or for getting a count of how many nodes are contained by a certain root.
This function will be used frequently.
CREATE FUNCTION get_contained_animals (integer)
RETURNS setof integer AS'
SELECT child FROM n_contains_a WHERE parent IN (get_leaf_nodes($1));
' language 'sql';
As mentioned previously, this system will involve a graphical web interface. This should greatly clarify its use. I imagine the main user activity will be browsing classifications, or searching directly for animal records (encyclopedia use). There is a question of how to display a classification tree. For lack of a better idea, a file-system-like structure could be shown, where the user moves through the tree one level at a time. This tree navigation would happen in one frame. Meanwhile, information about the current root, node, or animal would be displayed in another frame. Along the bottom of the screen would be a search interface frame. This would allow users to search at any time. Searches could be restricted to the set of animals under the current node or frame, or from an intersection of nodes chosen using the tree navigation frame. See the image below for a mockup of such an interface.
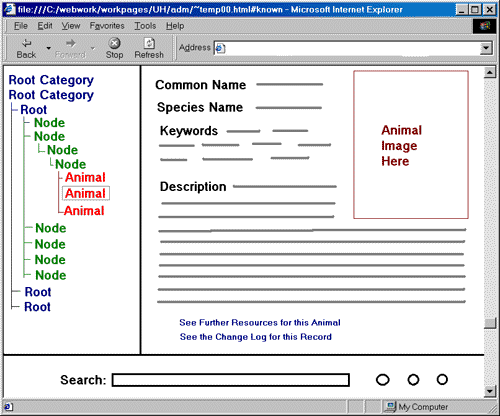
There are a number of advantages for using such an interface. Without it, it is certainly hard to see any structure; there are only rows and rows of "contains" relationships. It gives the user access to the system without requiring them to know SQL. It keeps track of certain information for the user. For example, the current TEid would be in a hidden field in the HTML of the current page; it is used in querying for information related to that entity, yet the user never encounters it. The interface also provides some security. It ensures at least some filtering of input, whether searches or data input. Through this filtering and structuring, malicious use can be restricted. For example, LOBS are returned one or two at a time--only when displaying the related animal record. By not allowing the user to query the DB directly, we need not worry about what happens when the user requests to all animal images in the database at once.
By using the interface described above, the SQL queries processed by the system are simplified, though frequent. Here are some examples.
Clicking on a node in the tree navigation frame displays the next layer of nodes beneath it. The interface passes along the selected node's TEid without the user's knowledge. Thus the interface would send the following SQL query to get the information it needs.
SELECT id, taxon_name FROM nodes
WHERE id IN (SELECT get_contained_nodes(TEid);
To see the resources for a record, whether animal, node, or root:
SELECT * FROM resources
WHERE (TEid = cites.teid) AND (cites.source = resources.id);
Or get this record's change log (which is essentially information need #7):
SELECT username, time FROM edited
WHERE TEid = edited.teid;
The interface does not prevent more complex queries, however. Here is the SELECT statement produced for information need #7 concerning blubberless arctic marine mammals.
SELECT id, common_name, species_name FROM animals
WHERE (id IN get_contained_animals(arctic TEid)
AND id IN get_contained_animals(marine TEid)
AND id IN get_contained_animals(mammals TEid))
AND description NOT ILIKE '%blubber%';
ILIKE is PostgreSQL's case-insensitive version of LIKE. This query could also use PostgreSQL's regular expressions, though doing so would probably be overkill here. To do exactly the same case-insensitive search with regex, the last line would instead read:
AND description !~* 'blubber';
While working on this project, I discovered a number of features that would be desirable in a OR-DMBS or in SQL. First of all, I would prefer a more object-oriented programming approach. I think what I long for is an application from Stonbraker's Quandrant 3! Perhaps because I am new to SQL, I have difficulty stating my needs without obvious control statements. I think it'd be easier with a few arrays and if statements.
Another thing that would be nice is more IR processing of text. This will be more important during the early stages of the system when "encyclopedia-like" use is high. Some sort of ranking or weighting for text searches would be desirable. Most text searches in this system will likely include both keywords and description fields. A ranking algorithm could give more weight to those terms in keywords, and also increase the relevancy ranking based on word count. A more extensible search would also be nice--phrases, wild cards, prefix and suffix, proximity, etc. However, most of these can be simulated with PostgreSQL's support of POSIX regular expressions. Because I doubt most of my users will be such sophisticated searchers, I would suggest that the web interface support wildcards, phrases, and prefix/suffix, and that these be implemented behind the scenes with regular expressions. Since these added features can be overkill, and depend on the nature of the CLOBs and text used, I would not want them to replace LIKE as the default SQL3 behavior. However, added standard functions would be pleasant. Yet, in this system, I think that emphasising the meta-data provided for each animal by the various roots and nodes that contain it is more advantageous that trying to generate a better ranking algorithm for the text of the descriptions.
One benefit of PostgreSQL is that is supports functions written in nearly any language (if you can figure out how to load the necessary module.) This allows programmers and implementers to write as much as they can in the languages they are used to. Great for UDFs. I'm not sure if other other vendors are as flexible.
What I think SQL needs most in the next version is consistency between implementations. It's hard to learn when even the "core" part of the language has subtle syntax and term variations between vendors.
Another issue we've discussed during this course is how to handle a large number of LOBs in search results. I think I've largely avoid that problem in this case with the web interface. Indeed, as databases attempt to reach more users, things will need to be simpler and more user-friendly. One aspect of that user-friendliness is not returning 1,000 images as the results of a query with no way for the user to organize them or scroll through them or even stop them if they are being transmitted across a network. Yet, on the other hand, there is the old debate about operating systems that is relevant here. People often point out that Unix has very few safety catches and user aids. But when you need power, it's what you use. In the same way, I think we should be wary of tampering SQL such that it decides for the user what is too much data. For this reason, I propose interfaces when possible; database implementers, who know the nature of the system, can exert some control on acceptable searches. If some control must be added directly to SQL, I would propose a confirmation option. If a user is about to receive an huge number of results or amount of data, the system might report the number of rows or objects or bytes about to be returned and get a confirmation before returning them. Different techniques for displaying or organizing results really depends on the data and the user and should probably not be standardized, though extra, optional functions or aids would be useful.
For more specific difficulties, see Appendix
Overall, I feel this has been a fruitful project. It has exposed me to a number of issues, both in design and SQL implementation I would never had encountered through reading alone. I also learned that a project such as this never ends. There are always further issues and problems to solve, new features to add. Yet I feel that implementing this system is not an impossible goal. Comments or questions are appreciated.
PostgreSQL Global Development Group. PostgreSQL 7.1 Documentation <http://www.ca.postgresql.org/users-lounge/docs/7.1/postgres/> Last Accessed: 16 Dec 2001.
Momjian, Bruce. PostgreSQL: Introduction and Concepts. <http://www.ca.postgresql.org/docs/aw_pgsql_book/index.html> Last Accessed: 16 Dec 2001.
Worsley, John, and Joshua Drake. Practical PostgreSQL. <http://www.postgresql.info/> Last Accessed: 16 Dec 2001.
TreeEntity:
roots, nodes, or animals. For this reason, I was forced to include the table_ref column of tree_entities. Pity that one can't query an OID for what table its in, since it unique between all tables.
Animal:
Root:
Resource:
User:
Creator:
contains:
opened:
| ~ztomasze Index:
LIS: Project Report http://www2.hawaii.edu/~ztomasze |
Last Edited: 16 Dec 2001 ©2001 by Z. Tomaszewski. |