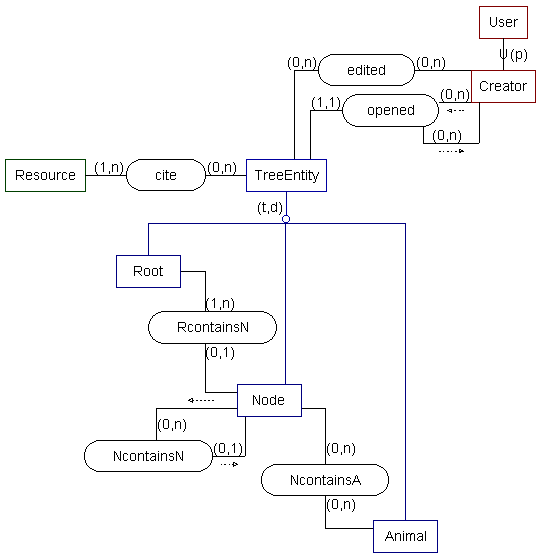
-- SSM model --
The following in a data model and beginning implementation plan for an animal classification system. This system contains multimedia records for many types of animal. The system allows for the construction of a variety of classifications by using Node objects. By adding new classifications or exploring existing ones, users can examine a variety of relationships between animals. They can use alternate classifications, or even intersect different classifications, depending on their current information need.
I imagine this system as a web-based, open-access system. It will be an animal encyclopedia with text and images of specific animal species. It can serve as a partial index to related works, especially since cited resources can be URLs as well as print materials. It will be a taxonomy modeling tool. Most importantly, the combination of multiple user-created classifications will make it a powerful searching and metadata system.
The system would probably be setup and maintained by either a volunteer group, non-profit organization, or university. There should be a graphical Web interface so that users can browse, see classification structures, and get the full benefit of the images and URL links to other sources. The system will be implemented using PostgreSQL.
Possible users include:
Users such as high school students and some amateur enthusiasts will likely use the system mainly as an encyclopedia to find more information on certain animals. Such uses do not involve building new classifications or adding animal records. These users will be Users in terms of the SSM below.
Other amatuer enthusiasts may compile information from various sources and create new animal records or edit current ones. Taxonomists may use the system to build models of their work. Biology researchers may add their findings to animal records. They may also use the system to examine possible relationships made visible through intersecting classficiations, such as effect of climate on gestation period. These users that create new records will be Creators in terms of the SSM below.
Here are some example information needs that illustrate the system.

-- SSM model --
| Animal | |
|---|---|
| id | <integer> -- a TreeEntitiy foreign key |
| species_name | <char(60)> -- the Latin Genus-species name for this animal |
| common_name | <char(60)> -- the common English name for this animal. |
| keywords | <vchar(200)> -- keywords describing this animal to aid in searching |
| Description | <text> |
| Image | filename: <char(30)> -- the name of the image file pixel-width: <integer> -- the width of the image in pixels pixel-height: <integer> -- the height of the image in pixels format: <char(4)> -- {JPEG | GIF | PNG | BMP} comments: <vchar(500)> -- any comments on the image image: <image> |
| Root | |
|---|---|
| id | <integer> -- a TreeEntitiy foreign key |
| classif_name | <char(60)> -- the name of the classification system stemming from this root |
| keywords | <vchar(200)> -- keywords describing this classification to aid in searching |
| scope_all | <boolean> -- whether this classification aims to include all animals (as opposed to just a subset) |
| description | <vchar(1000)> -- a description of this classification system |
| Node | |
|---|---|
| id | <integer> -- a TreeEntitiy foreign key |
| taxon_name | <char(60)> -- the name of the particular taxon or group or classification level represented by this node |
| type | <char(40)> -- describes the type of taxon or the level within the classification system |
| keywords | <vchar(200)> -- keywords describing this node to aid in searching |
| Description | <vchar(1000)> -- a description of this entity |
| TreeEntity | |
|---|---|
| id | <sequence> -- a unique system id for this entity |
| Resource | |
|---|---|
| id | <dec(12)> |
| author | <char(50)> |
| title | <char(200)> |
| publisher | <char(60)> |
| location_or_URL | <char(100)> |
| pub_date | <date> |
| notes | <vchar(250)> -- notes or comments about this resource |
| User | |
|---|---|
| username | <char(20)> -- a unique username |
| age | <integer> -- assumes that centenarians are not web-surfing taxonomists |
| education | <char(11)> {ELEMENTARY | MIDDLE | HIGH SCHOOL | COLLEGE | GRAD | TEACHER | PROFESSOR} |
| nationality | <char(30)> |
| <char(30)> | |
| created | <date> |
| last_accessed | <date> |
| time_inactive | <derived> -- how long it has been since this user logged in |
| Creator | |
|---|---|
| password | <char(12)> |
| name | <char(50)> |
| address | <char(120)> |
| release_prefs | <char(8)>{USERNAME | NAME | EMAIL | ADDRESS} -- how much information to release to other users; each choice includes those before it. |
| greeting | <vchar(750)> -- an introductory message to other users of the system |
| RcontainsN | |
|---|---|
| E1: Root | (1,n) -- A Root must contain some Nodes. |
| E2: Node | (0,1) -- A Node may or may not be contained by a Root |
| NcontainsN | |
|---|---|
| E1: Node | (0,n) -- A Node may or may not contain other Nodes. |
| E2: Node | (0,1) -- A Node may be contained by another Node. |
| NcontainsA | |
|---|---|
| E1: Node | (0,n) -- A Node may or may not contain Animals |
| E2: Animal | (0,n) -- An Animal may be contained by any number of Nodes, or none at all. |
| cite | |
|---|---|
| E1: TreeEntity | (0,n) -- An entity may cite any number of Resources. |
| E2: Resource | (1,n) -- A Resource must be cited at least once. |
| edited | |
|---|---|
| E1: Creator | (0,n) -- A Creator may edit any number of entities. |
| E2: TreeEntity | (0,n) -- An entity can be edited any number of times. |
| time | <timestamp> -- The time and date the new TreeEntity was edited. |
| opened | |
|---|---|
| E1: Creator | (0,n) -- A creator may not have created an entity yet; if he has, he can create any number of them |
| E2: TreeEntity | (1,1) -- Each set of access permissions is related to only a single Entity. |
| E3: Creators | (0,n) -- A Creator grants access to any number of Creators, 0 being all. |
| time | <timestamp> -- The time and date the new TreeEntity was added. |
For more information on the above entities and relationships, see v0.3. The examples there may also make the system easier to understand.
I forsee the need to create a table for each SSM entity and relationship above, plus a table for Image objects. Thus, I will have the following tables:
tree_entities
roots
nodes
animals
resources
users
creators
cited
edited
opened
r_contains_n
n_contains_n
n_contains_a
image
Here are some example CREATE statements from this system.
CREATE SEQUENCE teid_counter;
CREATE TABLE tree_entities (
teid INTEGER DEFAULT nextval('teid_counter') PRIMARY KEY,
entity_oid OID
);
CREATE TABLE roots (
id REFERENCES tree_entities PRIMARY KEY,
classif_name char(60),
keywords varchar(200),
scope_all boolean,
description varchar(1000)
);
CREATE TABLE r_contains_n (
root_id REFERENCES roots,
node_id REFERENCES nodes UNIQUE
);
I plan to index the following columns for faster retrieval (in order of imporance):
If possible, it would be nice to also form indices of keywords from the the content of all the keywords and descriptions fields, as well as from animal text objects.
Certain functions will also be required. They should do the following:
get_contained_animals(TEid)
get_contained_nodes(TEid)
Comments or questions are appreciated!
Thank you for reading this far! The following is a working list of notes, thoughts, and running commentary concerning current problems and future developments. Though not officially part of this report, the content of these appendices delimit the known problems . Thus comments on these points would be most helpful.
After reading more on SQL3, I have learned the GRANT, REVOKE NOTIFY, and LISTEN commands might be possible replacements to the opened and edited relationships. GRANT does require a sequence, but this could be (relatively) easily incorporated as the TreeEntity ID. However, I do not know if this access would be inherited from this Entity to Node, Root, and Animal, which is where it is actually needed.
Benefits of switching:
Drawbacks of switching:
This needs more study.
Update: After further study, I realized that GRANT applies to the sequence relation (a counter basically), and not to the rows numbered using that sequence. Thus, after all that, I realized I can not use GRANT to control access to the entity objects of the system because they will correspond to table rows and not distinct tables.
At first this seemed like a good idea, but after further consideration I realized it would make determining parents impossible. Admittedly, working up a tree will be much more rare, but it still seems a necessary feature.
Just as this page was going to "press" (this project is ongoing and doesn't seem to stop for progress reports) I realized that, while OIDs are unique identifiers for rows, I can't seem to use them without also knowing the table they're in. This was the reason I started using them in the first place; I do not yet know how to determine which table (roots, nodes, or animals) a certain entity (row) is in. I suppose could just SELECT FROM all three tables; because the teid is unique, the id will occur in only one of the tables. The other alternative is to include a table-identifying column in tree_entities.
Animal:
Root:
Resource:
User:
contains:
opened:
See v0.3's Wishlist.
| ~ztomasze Index:
LIS: Initial Data Model http://www2.hawaii.edu/~ztomasze |
Last Edited: 30 Oct 2001 ©2001 by Z. Tomaszewski. |