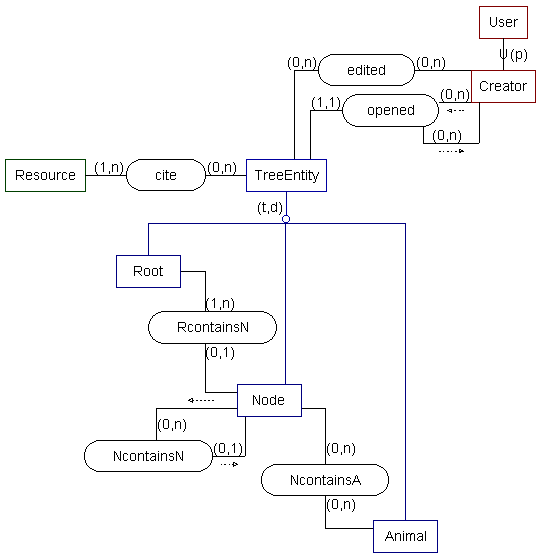
-- SSM model --
The following system contains multimedia records for many types of animal. The system allows for the construction of a variety of classifications by using Node objects. By adding new classifications or exploring existing ones, users can examine a variety of relationships between animals. They can use alternate classifications, or even intersect different classifications, depending on their current information need. See v0.4 for more information.
Here are some example information needs that illustrate the system.
This is a relatively simple and common query for the system: return all the images of animals under this node. However, it demonstrates how this information comes from multiple tables and how user-defined functions will play a large role.
The following tables will be involved:
animals
images
tree_entities
nodes
n_contains_n
n_contains_a
As well as the function:
get_contained_animals(TEid)
This statement assumes that user already knows the TEid for the Arctic node. It should return the common name, species name, and image of every animal contained by this Arctic node.
SELECT animals.common_name, animals.species_name, images.image
FROM images, animals
INNER JOIN animals ON (animals.teid = images.animal_id)
WHERE images.animal_id = get_contained_animals(node TEid);
This query uses a user-defined function. As such, the DBMS should include such extensions as user-defined selectivity functions, smart ordering of clauses, and optimization of expensive functions.
If there is no way to tell the system which to do first, then hopefully the WHERE clause is performed before the JOIN. If the JOIN is performed first, there could be up to a million rows joined (the number of known species). If it is performed after the WHERE, only tens or possibly hundreds of rows will be joined (only arctic animal species).
For my proposed system, it is important that the DBMS be able to handle large tables and access them quickly. Most of the frequently used and important information is present in the relationships between entities, rather than only as data within those entities. As such, TEid columns in all tables are frequently compared, especially by the recursive "get child entity" functions. Hopefully the system will handle such tasks quickly. This does not seem too high of a demand since TEids are easy to sort and compare.
Since OR-DBMSs must allow UDTs and UDFs, it is important for them to make "educated guesses" on the most efficient order of processing queries. This task is much more difficult and involved than in relational databases because the details of the functions and task times are not known by the OR-DBMS, as they are in a traditional relational system.
Comments or questions are appreciated.

-- SSM model --
| Animal | |
|---|---|
| id | <integer> -- a TreeEntitiy foreign key |
| species_name | <char(60)> -- the Latin Genus-species name for this animal |
| common_name | <char(60)> -- the common English name for this animal. |
| keywords | <vchar(200)> -- keywords describing this animal to aid in searching |
| Description | <text> |
| Image | filename: <char(30)> -- the name of the image file pixel-width: <integer> -- the width of the image in pixels pixel-height: <integer> -- the height of the image in pixels format: <char(4)> -- {JPEG | GIF | PNG | BMP} comments: <vchar(500)> -- any comments on the image image: <image> |
| Node | |
|---|---|
| id | <integer> -- a TreeEntitiy foreign key |
| taxon_name | <char(60)> -- the name of the particular taxon or group or classification level represented by this node |
| type | <char(40)> -- describes the type of taxon or the level within the classification system |
| keywords | <vchar(200)> -- keywords describing this node to aid in searching |
| Description | <vchar(1000)> -- a description of this entity |
| TreeEntity | |
|---|---|
| id | <sequence> -- a unique system id for this entity |
| NcontainsN | |
|---|---|
| E1: Node | (0,n) -- A Node may or may not contain other Nodes. |
| E2: Node | (0,1) -- A Node may be contained by another Node. |
| NcontainsA | |
|---|---|
| E1: Node | (0,n) -- A Node may or may not contain Animals |
| E2: Animal | (0,n) -- An Animal may be contained by any number of Nodes, or none at all. |
| ~ztomasze Index:
LIS: Initial Data Model http://www2.hawaii.edu/~ztomasze |
Last Edited: 21 Nov 2001 ©2001 by Z. Tomaszewski. |